Selected first/senior author publications
Click here for All Publications
(Total: 72 | First author: 7 research articles | Senior author: 44 research articles & reviews)
[** - (Co-)Corresponding author]
2026
Chung CH, Bhowmick R, Badenoch AJ, Arora HS, Chandrasekaran S**, Targeting metabolism to combat anticancer and antibacterial drug resistance, Trends in Pharmacology (in press).
Arora, H. S., Lev, K., Robida, A., Velmurugan, R., & Chandrasekaran, S**. A mechanistic neural network model predicts both potency and toxicity of antimicrobial combination therapies. NPJ Drug Discovery (in press).
2025
Pallikkavaliyaveetil N, and Chandrasekaran S**, Small data, big challenges: Machine- and deep-learning strategies for data-limited drug discovery, Advanced Drug Delivery Reviews.
Woodard, J.C., Liu, Z., Chegini, A.M., Tian, J., Bhowmick, R., Pennathur, S., Mashaghi, A., Brender, J. and Chandrasekaran, S**. A structural machine learning approach for rapid prediction of thermodynamically destabilizing tyrosine phosphorylations. Cell Reports Methods.
Lin D, Zhang L, Zhang J, Chandrasekaran S**, Inferring Metabolic Objectives and Tradeoffs in Single Cells During Embryogenesis, Cell Systems. Press release: Unlocking Cellular Objectives Through Machine Learning.
Inferring metabolic objectives and tradeoffs with SCOOTI
2024
Schildcrout, R., Smith, K., Bhowmick, R., Menon, S., Kapadia, M., Kurtz, E., Coffeen-Vandeven A, & Chandrasekaran S**. Recon8D: A metabolic regulome network from oct-omics and machine learning. BioRxiv.
Chung C, Chang D, Rhoads N, Shay M, Srinivasan K, Okezue M, Brunaugh A, Chandrasekaran S**, Transfer learning predicts species-specific drug interactions in emerging pathogens, BioRxiv.
Bhowmick R., Campit S., Keshamouni V. G., & Chandrasekaran S**. Constraint-based modeling identifies metabolic vulnerabilities during the epithelial to mesenchymal transition. Communications Biology.
Sambarey A, Smith K, Chung CH, Arora HS, Yang Z, Agarwal P, Chandrasekaran S**, Integrative analysis of multimodal patient data identifies personalized predictors of tuberculosis treatment prognosis, iScience. Press release: Multimodal AI to guide personalized treatments for TB. Highlighted by The Conversation, SFGate, Houston Chronicle, Deccan Herald & UM President Ono
Using Multimodal AI to Guide Tuberculosis Treatment
Eames A, Chandrasekaran S**, Leveraging metabolic modeling and machine learning to uncover modulators of quiescence depth, PNAS Nexus. Press release: BME grad uses AI to predict Quiescence
Quiescence depth prediction using machine learning and genome-scale metabolic modeling
2023
Scott Campit, Rupa Bhowmick, Taoan Lu, Aaditi Saoji, Ran Jin, Aaron Robida, Chandrasekaran S**, Data-Driven Screening to Infer Metabolic Modulators of the Cancer Epigenome. bioRxiv [preprint]
Mohammad Askandar Iqbal**, Shumaila Siddiqui, Kirk Smith, Prithvi Singh, Bhupender Kumar, Salem Chouaib, and Chandrasekaran S**, Pathway-Based Analysis of Breast Tumors Reveal Metabolic Subtypes of Clinical Relevance. iScience.
2022
Chung, C. H., & Chandrasekaran, S**. A flux-based machine learning model to simulate the impact of pathogen metabolic heterogeneity on drug interactions. PNAS Nexus.
CARAMeL combines machine learning with metabolic modeling to predict how drug interactions are impacted by both extrinsic and intrinsic metabolic heterogeneity
Cantrell J, Chung CH, Chandrasekaran S**, Machine learning to design antimicrobial combination therapies: promises and pitfalls, Drug Discovery Today [review article].
2021
King J, Patel M, Chandrasekaran S**. Metabolism, HDACs, and HDAC Inhibitors: A Systems Biology Perspective. Metabolites [review article].
Lee HJ**, Shen F, Eames A, Jedrychowski MP, Chandrasekaran S**. Dynamic metabolic network modeling of a mammalian cell cycle using time-course multi-omics data, bioRxiv. [Preprint]
Smith K, Shen F, Lee HJ, Chandrasekaran S**. Metabolic signatures of regulation by phosphorylation and acetylation. iScience. [Download CAROM]
Predicting PTM regulation using CAROM
Chung CH, Lin D-W, Eames A, Chandrasekaran S**. Next-Generation Genome-Scale Metabolic Modeling through Integration of Regulatory Mechanisms. Metabolites. [review article].
Overview of Next-generation GEMs that incorporate multiple regulatory mechanisms
Chandrasekaran S**, Danos N, George UZ, Han JP, Quon G, Müller R, Tsang Y, Wolgemuth C, “The Axes of Life: A roadmap for understanding dynamic multiscale systems”, Integrative and Comparative Biology. [review article] [Editor’s Choice]
Cicchese J, Sambarey A, Kirschner DE, Linderman JJ**, Chandrasekaran S**. "A multi-scale pipeline linking drug transcriptomics with pharmacokinetics predicts in vivo interactions of tuberculosis drugs." Scientific Reports.
Simulating efficacy of Tuberculosis drug regimens at the molecular, cellular and granuloma scale
2020
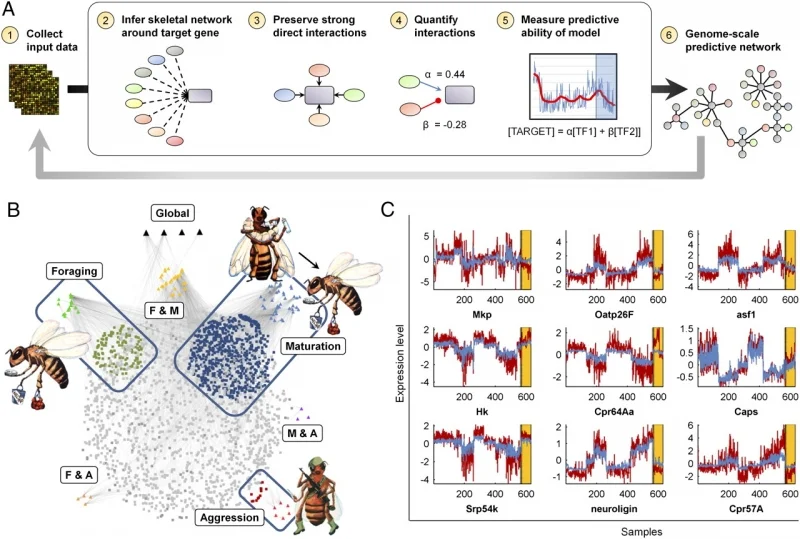
Jones, BM, Rao VD, Gernat T, Jagla T, Cash-Ahmed A, Rubin BE, Middendorf M, Sinha S, Chandrasekaran S**, Robinson GE**. “Individual differences in honey bee (Apis mellifera) behavior enabled by plasticity in brain gene regulatory networks”. eLife. Press release: Brain gene expression patterns predict behavior of individual honey bees
Predicting regulators of bee behavior using AI
Campit S, Meliki A, Youngson N, Chandrasekaran S**, Nutrient Sensing by Histone Marks: Reading the Metabolic Histone Code Using Tracing, Omics, and Modeling. BioEssays. [review article]
Studying metabolic-epigenetic interactions using systems biology
Oruganty K, Campit S, Mamde S, Lyssiotis C, Chandrasekaran S**, Common biochemical properties of metabolic genes recurrently dysregulated in tumors, Cancer and Metabolism.
Campit S and Chandrasekaran S**, Inferring Metabolic Flux from Time-Course Metabolomics, Methods in Molecular Biology: Metabolic Flux Analysis in Eukaryotic Cells, [Preprint PDF, Supplement]. [review article]
Chandrasekaran S, Deep learning AI discovers surprising new antibiotics, The Conversation. [magazine article]
Harnessing AI to find antibiotics against pathogens like MRSA (Source: NIAID)
2019
Ma S, Jaipalli J, Larkins-Ford J, Lohmiller J, Aldridge B, Sherman D**, and Chandrasekaran S**, Transcriptomic signatures predict regulators of drug synergy and clinical regimen efficacy against Tuberculosis, mBio. Summary: Designing new TB drugs using AI. Press release: How an AI solution designs new tuberculosis drug regimens. (** - Corresponding authors)
Predicting both drug interactions and regulators of drug interactions using INDIGO
Chandrasekaran S. Tying Metabolic Branches With Histone Tails Using Systems Biology. Epigenetics insights. [review article]
Shen F, Boccuto L, Pauly R, Srikanth S, Chandrasekaran S**, Genome-scale network model of metabolism and histone acetylation reveals metabolic dependencies of histone deacetylase inhibitors, Genome Biology. (Download Metabolism-Epigenome model). Summary: How metabolism affects gene activity via the ‘Histone code’.
Network model of cancer metabolism and histone acetylation
Shen F, Cheek C, & Chandrasekaran S**. Dynamic Network Modeling of Stem Cell Metabolism, Methods in Molecular Biology: Computational Stem Cell Biology. [Download protocol and Preprint]. [review article]
Chandrasekaran S, A Protocol for the Construction and Curation of Genome-Scale Integrated Metabolic and Regulatory Network Models, Methods in Molecular Biology: Microbial Metabolic Engineering.[Preprint PDF]. [review article]
2018
Cokol M**, Li C and Chandrasekaran S**, Chemogenomic model identifies synergistic drug combinations robust to the pathogen microenvironment, PLOS Computational Biology. (Download MAGENTA). Press release: A ‘decathlon’ for antibiotics puts them through more realistic testing.
Overview of MAGENTA to predict the impact of environment on drug interactions
Chandrasekaran S, Predicting drug interactions from chemogenomics using INDIGO, Methods in Molecular Biology: Systems Chemical Biology, [Preprint PDF]. [review article]
2017
Dotiwala F, Santara SS, Binker-Cosen A, Li B, Chandrasekaran S**, Lieberman J**, “Granzyme B disrupts central metabolism and protein synthesis in bacteria to promote an immune cell death program”, Cell (** - Corresponding author). Featured in Nature Reviews Microbiology and Cell Systems. Press release - Closest look yet at killer T-cell activity could yield new approach to tackling antibiotic resistance
Metabolic pathways targeted by granzyme-B
Chandrasekaran S*, Zhang J*, Ross C, Huang Y, Asara J.M., Li H, Daley G.Q., Collins J.J. "Comprehensive mapping of pluripotent stem cell metabolism using dynamic genome-scale network modeling", Cell Reports. Featured in Cell Systems. [Download source code and tutorial for the dynamic metabolic modeling approach used in this study]
2016
Chandrasekaran S**, Cokol-Cakmak M, Sahin N, Yilancioglu K, Kazan H, Collins J.J and Cokol M**, “Chemogenomics and Orthology-based Design of Antibiotic Combination Therapies”, Molecular Systems Biology (Cover Article). Featured in MedChemNet. (Download INDIGO here) (** - Corresponding author)
2010-2015
Chandrasekaran S*, Rittschof C*, Djukovic D, Gu H, Raftery D, Price N.D, and Robinson G.E, “Aggression is Associated with Aerobic Glycolysis in the Honey Bee Brain”, Genes Brain and Behavior, 2015.(Top 5 most cited articles in 2015 in Genes Brain and Behavior)
Chandrasekaran S, “Predicting Phenotype from Genotype through Reconstruction and Integrative Modeling of Metabolic and Regulatory Networks”, Systems and Synthetic Biology: A Systematic Approach (ed V.V.Kulkarni/G.B.Stan/K.S.Raman), 2014 [review article]
Chandrasekaran S, “Transcriptional Regulation Of Metabolism And Behavior: Insights From Reconstruction And Modeling Of Complex Biochemical Networks” (dissertation), 2013
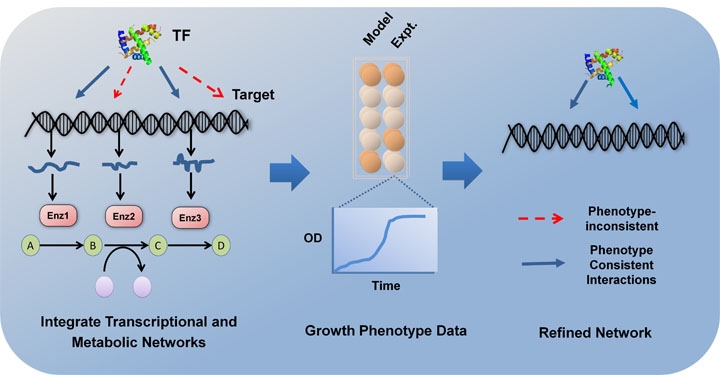
Chandrasekaran S and N.D. Price, “Metabolic Constraint-based Refinement of Transcriptional Regulatory Networks”, PLOS Computational Biology, 2013. Highlighted in Nature Genetics and Genomeweb.
Chandrasekaran S, Ament S.A, Eddy J.A, Rodriguez-Zas S.R, Schatz B.R, Price N.D, and Robinson G.E, "Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states", PNAS, 2011 (Cover Article). Highlighted in PNAS "Systems biology meets behavior". [Download ASTRIX algorithm and Supplementary datasets]
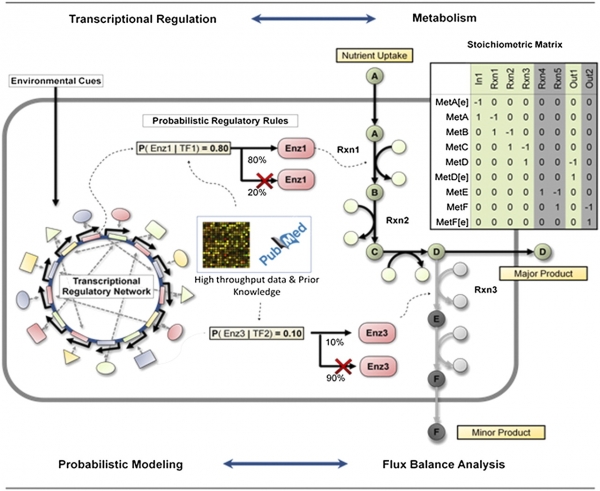
Chandrasekaran S and N.D. Price, "Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis," PNAS, 2010. [Download PROM models].